金桔
金币
威望
贡献
回帖0
精华
在线时间 小时
|
登陆有奖并可浏览互动!
您需要 登录 才可以下载或查看,没有账号?立即注册


×
tsRNA、rsRNA、piRNA这些非编码小RNA(sncRNA),可谓是液体活检领域冉冉升起的新星,我们之前也给大家介绍过潘多拉测序与非编码小RNA标志物开发研究方案(点击阅读)。在这篇文章,我们继续通过更多的案例,来更加深入的了解潘多拉测序在sncRNA疾病标志物研究上的运用。
sncRNAs
非编码小RNA (small non-coding RNAs, sncRNAs)是一大类长度较短且具有细胞调节功能的RNA的统称,在复制、转录和转录后调节因子以及染色质修饰复合物的向导方面发挥了关键作用[1,2]。非编码RNA在疾病机制中的作用也正在被发现,并且已经确定了几种ncRNAs作为潜在的药物靶标[3]。以往研究发现与癌症、心脑血管系统疾病、神经系统疾病、代谢性疾病和免疫系统疾病相关的突变约90%发生在非编码区,但是致病机理不明[4,5]。因此,对非编码小RNA的研究是非常有必要的。以往研究已对哺乳动物长度为19-24nt的miRNA,以及生殖细胞长度为20nt的特异表达的piRNA有着广泛和深入的认识,已成为表征最好的sncRNA类型,但迄今为止的持续探索已经确定了具有不同性质、生物发生途径、末端形成和分子功能的进一步功能性sncRNA[1]。
snoRNA
在上一篇文章中我们已经给大家介绍过tsRNA、piRNA、rsRNA,这次我们来讲讲snoRNA。小核仁RNA(small nucleolar RNAs, snoRNA) 是最早在核仁发现的一类中等长度的非编码小RNA,长度在60-300nt左右,大多数小核仁RNA可以分为C/D box snoRNA、H/ACA box两类[6,7,8]。snoRNA能与特定的蛋白质结合形成小核仁核糖核蛋白颗粒(small nucleolar ribonucleoprotein particle,snoRNP),参与rRNA的2’-O-甲基化或假尿苷化[9]。在脊椎动物中,编码snoRNA的基因主要存在于蛋白编码基因或非蛋白编码基因的内含子区域,并经过转录后加工产生成熟的snoRNA[10]。snoRNA最初的生物学功能是用来修饰rRNA,研究发现维持白血病干细胞活性的关键因素是SNORD和其指导的rRNA甲基化修饰[11]。除了进行rRNA的加工处理之外,还可以参与RNA剪接、翻译过程的调控以及氧化应激反应。研究表明snoRNA可以参与到遗传性疾病、代谢以及癌症的发生过程中。通过高通量测序技术测定人、黑猩猩、恒河猴和小鼠大脑前额叶皮质组织中snRNA与snoRNA的表达量,发现在人脑中的U1和SNORA29表达量发生了巨大的变化。其中,SNORA29 在人脑中的表达缺失,而在非人灵长类的神经元中特异表达,揭示了其在人脑认知功能形成中的潜在作用[12]。RNA-seq已成为研究RNA表达谱及其在正常生理和疾病过程的常用手段,该测序技术用于microRNA (miRNA)分析,极大的促进了非小编码RNA(sncRNA)的发现,但是标准RNA-seq仅捕获具有5 ' -磷酸(5 ' -P)和3 ' -羟基(3 ' -OH)末端的sncRNA,目前广泛使用的传统小RNA测序建库流程中包含的缺陷会导致测序结果产生系统性的偏差(bias),其它末端结构的sncRNAs不能通过标准RNA-seq有效扩增和测序,这是由于RNA修饰干扰了接头连接和逆转录过程,从而导致含有这些特殊修饰的小RNA无法在测序中被检测到[13,14]。形象地讲,这群小RNA就像披了一件隐形的斗篷(invisible cloak)。这些小RNA到底有多少?有什么重要的功能?这些都是学界尚未解决的问题。然而,随着这些sncRNA发挥的功能作用越来越重要,已展开研究适用于各种末端结构的sncRNAs测序技术[14]。
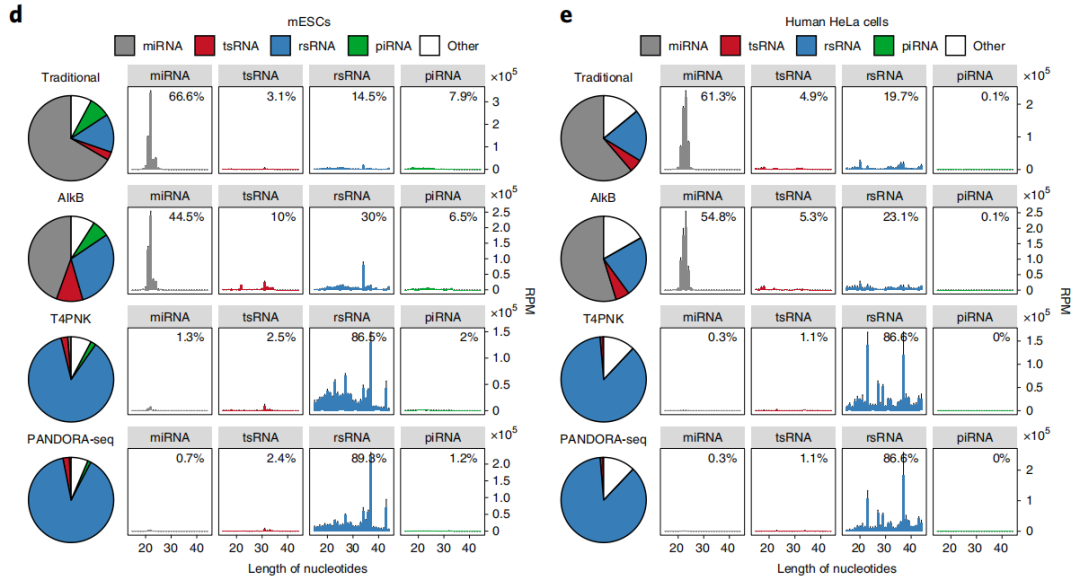
图1. 适用于各种末端结构的sncRNAs测序技术( 蓝色代表能够检测到的各种末端结构的sncRNAs)
PANDORA-seq
为了克服小RNA建库中的问题从而系统地发掘这些带有特定RNA修饰的小RNA,2021年4月5日,陈琦教授研究组在Nature Cell Biology杂志上发表了文章PANDORA-seq expands the repertoire of regulatory small RNAs by over-coming RNA modifications,建立了一套新的小RNA测序及分析流程PANDORA-seq,利用T4PNK 和AlkB对15-50nt区段的小RNA进行酶学处理,以去甲基化RNA修饰(例如,m1G, m1A, m3C和m22G)和促进接头连接的方式(将3’-P或者2’3’-cP 转换为3’-OH; 并添加5’-P)实现逆转录,解决了这些修饰妨碍逆转录酶通过的问题。PANDORA-seq优于传统测序以及使用单个AlkB或T4PNK处理后的测序结果,能够更广泛、更准确的在人类和小鼠组织、细胞中发现先前未知的含有特殊修饰的sncRNA。新方法鉴定了修饰sncRNAs主要是转移RNA来源的小RNA(tsRNAs)和核糖体RNA来源的小RNA(rsRNAs),这些以前未被检测到,在小鼠大脑、肝脏、脾脏和成熟精子中表现出组织特异性表达,以及在小鼠胚胎干细胞(mESCs)和人的HeLa细胞中表现出细胞特异性表达。PANDORA-seq揭示了体细胞向诱导多能干细胞(iPSCs)生成过程中前所未有的miRNA、tsRNA和rsRNA动态,以探究在胚胎干细胞(ESC)分化过程中的功能[14]。
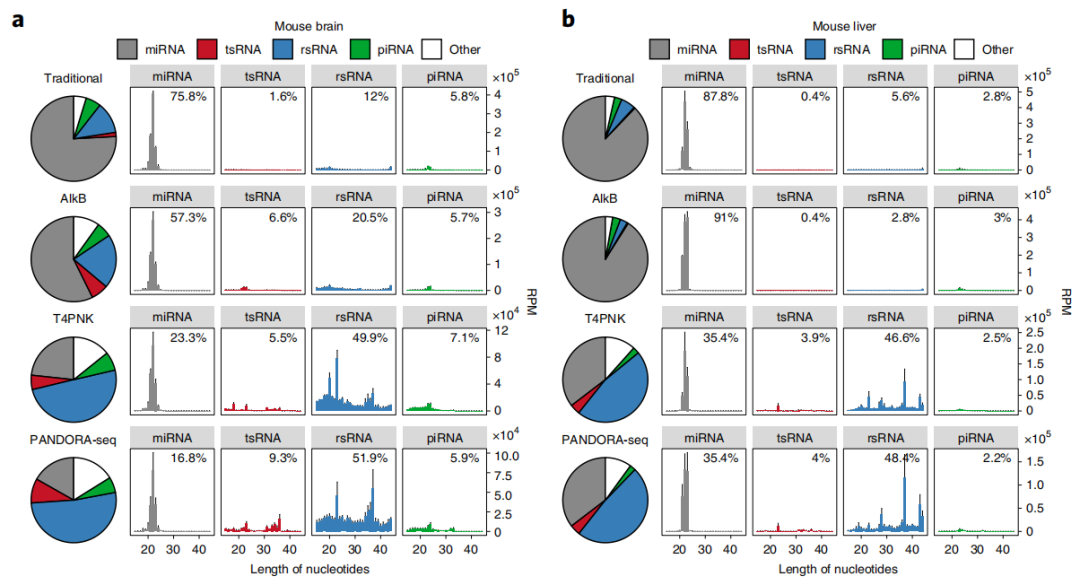

图2. 小鼠脑组织、小鼠肝脏、小鼠成熟精子、小鼠胚胎干细胞和人的HeLa细胞的小RNA测序结果(Traditional vs PANDORA-seq)
疾病sncRNA分子诊断标志物和功能机制研究方案
一、sncRNA分子疾病标志物筛选利用PANDORA-seq测序,揭示非编码小RNA(miRNA、piRNA、tsRNA、rsRNA、snoRNA)的差异表达谱,通过qPCR验证组织中的非编码小RNA,筛选跟疾病相关的发挥关键作用的小RNA分子;进一步通过化学合成sncRNAs模拟物mimics或者sncRNAs inhibitor转染细胞,并结合mRNA-seq测序进行差异分析以及IPA经典通路分析,筛选与疾病相关的sncRNAs,获得阳性表型sncRNAs,并构建诊断预后模型,筛选鉴定与疾病相关的诊断分子标记物;通过机器学习构建诊断预后模型,通过在训练集、验证集和测试集上反映预测效果,筛选鉴定分子诊断标记物;通过生物信息学方法预测目标sncRNAs靶基因,进一步结合WB、RIP-seq、RNA FISH、免疫荧光和双荧光素酶检测等实验研究sncRNAs对下游靶基因的调控作用;结合Ribo-seq测序,研究sncRNAs对机体整体翻译水平的影响,通过影响核糖体的生物发生增强翻译,与核糖体结合抑制翻译,或调控翻译起始过程抑制复合物形成,从而对处理组和对照组组织或细胞基因整体翻译水平变化进行研究,鉴定显著差异翻译效率的基因,绘制翻译图谱,这为深入理解sncRNAs在相关疾病发生过程中的功能机制和临床潜在的用药靶标提供新的指导思路。
二、实验验证及功能机制探究1)疾病组织差异小RNA筛选与验证(主要目标tsRNA)进行qPCR验证测序分析中得到的目的序列的表达量,为功能机制研究提供可靠数据;2)sncRNAs靶基因预测通过生物信息学方法,预测目标sncRNAs靶基因,对感兴趣的基因可以通过qPCR、WB和免疫组化法检测其在疾病组织的表达(筛选有生物学作用的RNA进行分子机制研究); 3)sncRNAs功能研究在疾病组织或细胞株中(选4~5种细胞株)检测sncRNAs的表达,使用sncRNAs mimic/inhibitor转染细胞,进行mRNA-seq测序,每组3个重复,进行差异分析和IPA经典通路分析,研究其对疾病组织或细胞增殖、侵袭及转移的影响 (可以根据靶基因的不同增加EMT、血管生成等指标的检测);4)sncRNAs对靶基因的表达调控机制研究通过WB,RIP,RNA FISH,免疫荧光,双荧光素酶等实验验证sncRNAs对靶基因表达的调控作用;并通过实验证实sncRNAs通过靶向目标基因发挥生物学作用。结合Ribo-seq测序研究sncRNAs对细胞基因翻译和翻译复合物形成的影响,发现sncRNAs参与翻译起始抑制、调节核糖体发生等细胞代谢过程的功能机制,进一步揭示sncRNAs在疾病的发生、发展过程中的生物学功能与潜在应用。
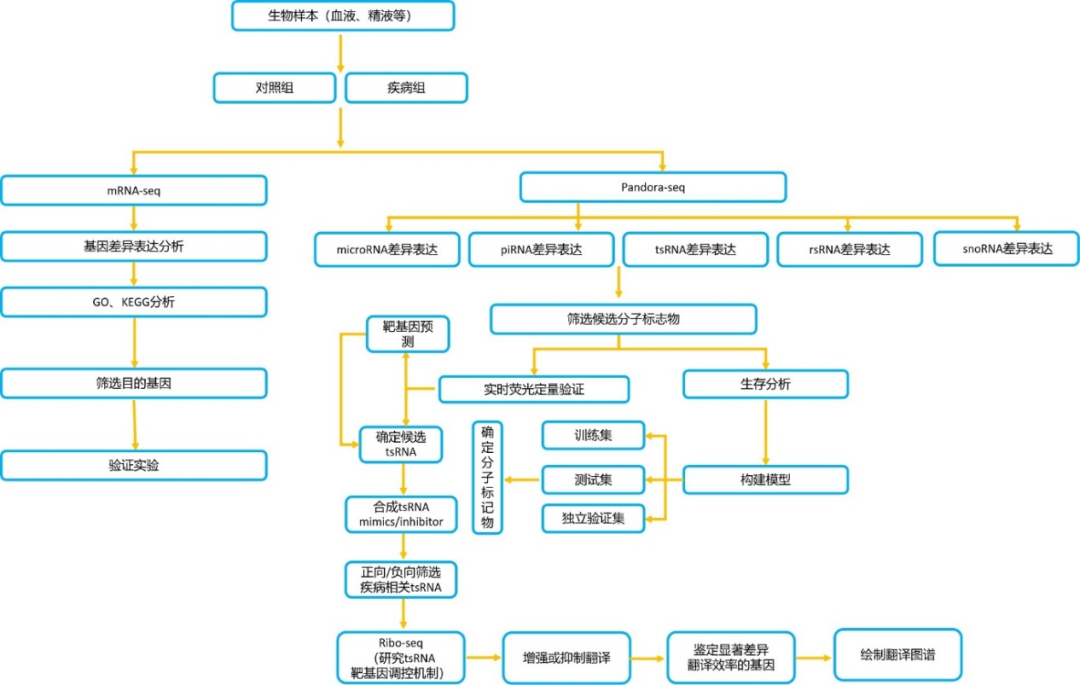
图3.疾病sncRNA分子诊断标志物和功能机制研究路径图(点击查看详图)
研究案例一
Paternal phthalate exposure-elicited offspring metabolic disorders are associated with altered sperm small RNAs in mice[15]
中文题目:父系接触邻苯二甲酸酯引起的后代代谢紊乱与小鼠精子小 RNA 的改变有关;
发表时间:2023-01-23
发表期刊:Environmental Science
影响因子:13.352
DOI:10.1016/j.envint.2023.107769
邻苯二甲酸酯是一种使塑料更耐用的化学品,邻苯二甲酸二环己酯(Dicyclohexyl Phthalate,DCHP)对小鼠F1、F2代代谢健康有着较大的影响。本研究通过将暴露于DCHP的F1雄性小鼠与未暴露的雌性小鼠繁殖产生F2代,利用PANDORA-seq方法发现接触DCHP后会导致精子中的小分子RNA发生变化,相较于传统的RNA测序方法,PANDORA-seq可以检测到更多变化,从而发现与疾病相关的关键调控的小分子RNA。先前研究报道了成熟的小鼠精子中的tsRNA和rsRNA含量较高,并且RNA修饰会改变二级sncRNA结构以及生物学特性。本研究结果表明DCHP处理后的雄性小鼠对后代代谢健康的不利影响,并可以诱导F1后代代谢紊乱,比如葡萄糖耐受不良,亲本雄性小鼠的代谢表型可以由F1 代传递给F2代,然而仅雌性F2后代中发现了这些紊乱。研究揭示了父系DCHP处理后可导致后代代谢健康的性别特异性跨代影响。
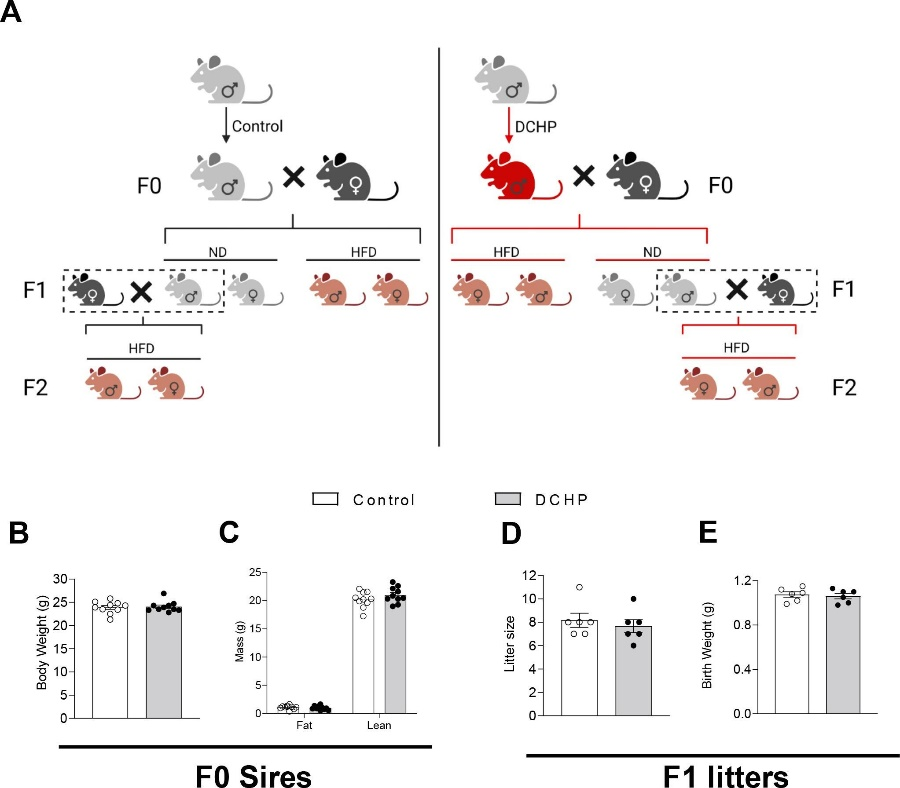
图4. 父系DCHP 暴露研究和DCHP 暴露对F0代和F1代的影响
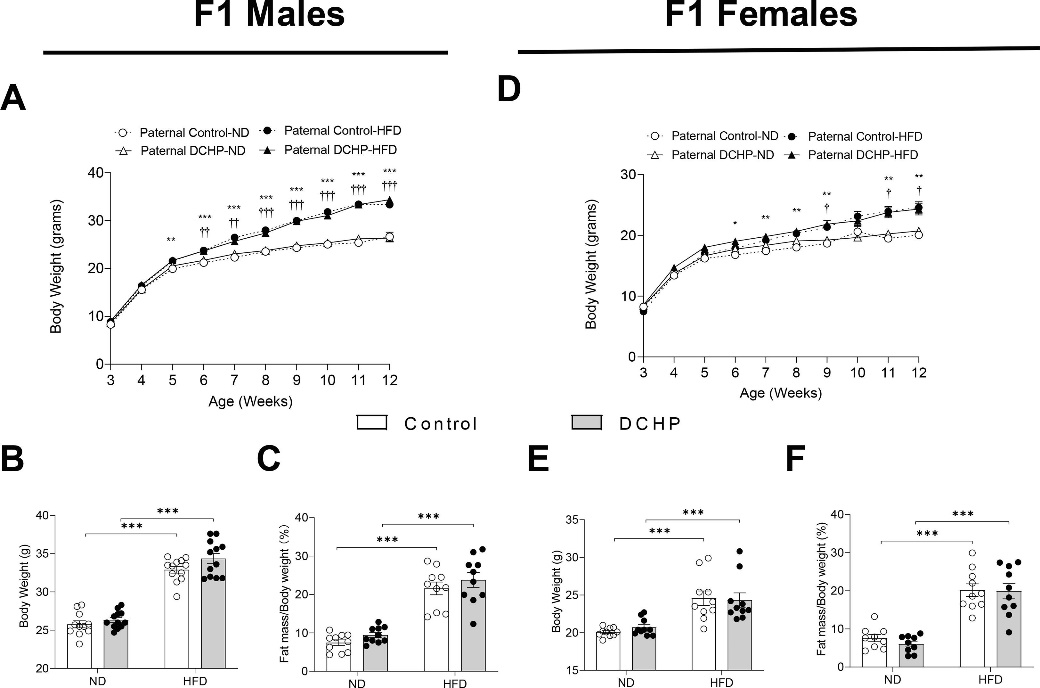
图5. 父系DCHP 暴露不会影响F1后代饮食诱导的肥胖
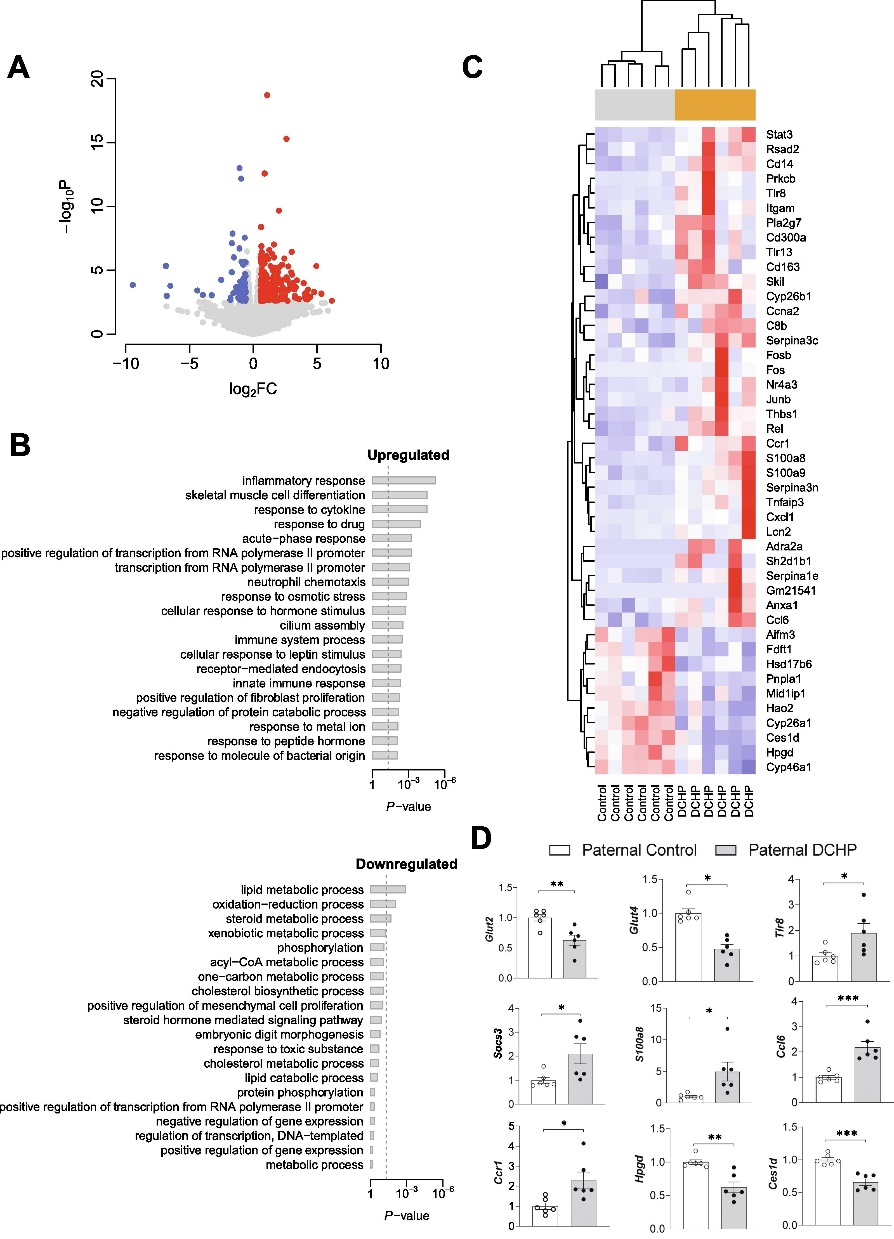
图6. 父系DCHP暴露改变F1雄性后代的肝脏转录组
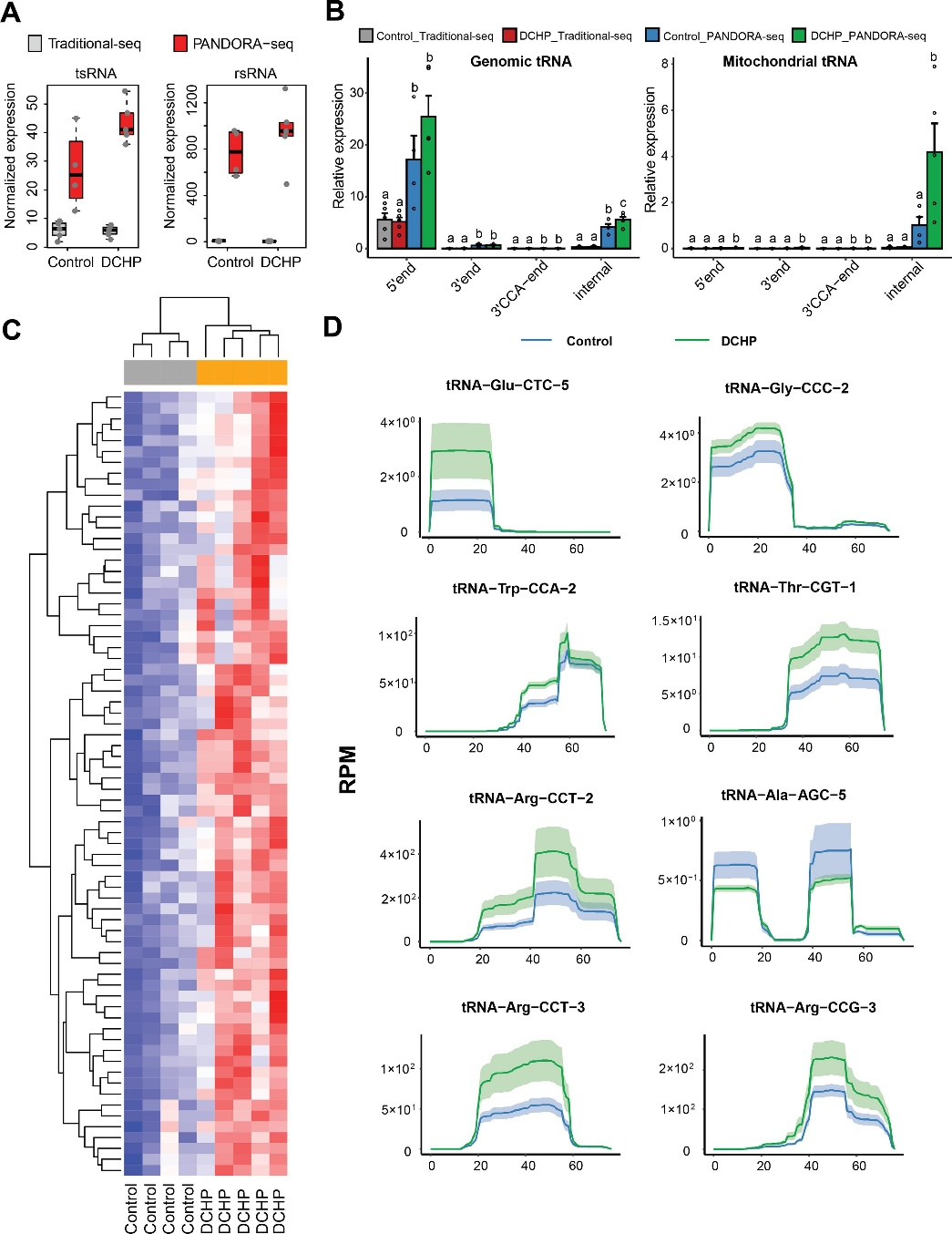
图7. PANDORA-seq接触DCHP后精子中识别出显着变化的 tsRNA 和 rsRNA
研究案例二
A transfer-RNA-derived small RNA regulates ribosome biogenesis[16]
中文题目:tRNA衍生的小RNA调控核糖体的生物发生
发表时间:2017-11-29
发表期刊:Nature
影响因子:69.504
DOI:10.1038/nature25005
tRNA衍生的小RNA(tsRNAs,tRNA-derived fragments)是一类丰富的非编码小RNA,其生物学作用尚不清楚。基于此,研究者从HeLa和HCT-116细胞的表达谱中筛选鉴定了tsRNA,利用LNA和ASO的混合物抑制多个基因,发现抑制LeuCAG3′tsRNA可以显著影响细胞活性。接着,进行了tsRNA测序,因为有丰度的翻译后tRNA修饰,所以常规测序技术不能准确定量tsRNA,所以这项研究还结合了Northern blot来鉴定tsRNA,发现主要的LeuCAG3′ tsRNA异构体应该是22nt而不是18nt。为了研究LeuCAG3‘tsRNA与翻译之间可能存在的关系,通过多聚核糖体分析(蔗糖密度梯度离心分析)发现LeuCAG3‘tsRNA被抑制后,40S和80S核糖体复合物减少,60S亚基的丰度增加。利用嘌呤霉素将80S和多聚核糖体解离为游离的40S和60S亚基,表明LeuCAG3‘tsRNA被抑制的细胞中40S和60S亚基的数量减少。与40S核糖体亚基的减少一致,抑制LeuCAG3‘tsRNA后,18S成熟rRNA的丰度减少。为了探究rRNA转录或加工是否受到LeuCAG3‘tsRNA抑制的影响,作者通过Northern定量研究了三种细胞系中不同的前体和成熟rRNA的丰度。抑制LeuCAG3‘tsRNA后,30S rRNA前体累积增加,而21S和18S rRNA前体减少。此外,作者使用小的抑制性RNA(siRNAs) 降低了这些蛋白质或大的核糖体蛋白RPL7的浓度,或XRN2或c-MYC的浓度,这些蛋白质对rRNA的生物发生很重要。然而,32S前rRNA和45S初级转录信号仅略有增加。综上所述,这些结果表明LeuCAG3‘tsRNA既没有显著影响28S成熟rRNA的加工,也不影响45S初级转录本的丰度,而是损伤了30S中间的5’端ETS(外转录间隔区)的去除,从而减少18S rRNA。本研究发现了LeuCAG3 tsRNA至少与两个核糖体蛋白mRNAs(RPS28和RPS15)结合以促进蛋白翻译。研究数据揭示了一种转录后机制,即可以微调不同生理状态下的基因表达,为治疗癌症提供一个潜在的新靶点。
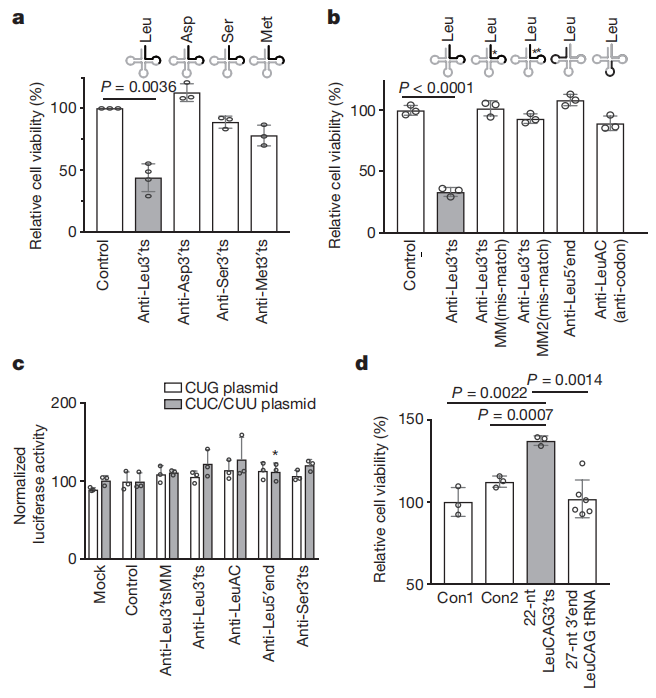
图8. LeuCAG3’tsRNA是细胞存活所必需的

图9. 抑制LeuCAG3’tsRNA诱导细胞凋亡,抑制PDX小鼠模型肝癌细胞的生长
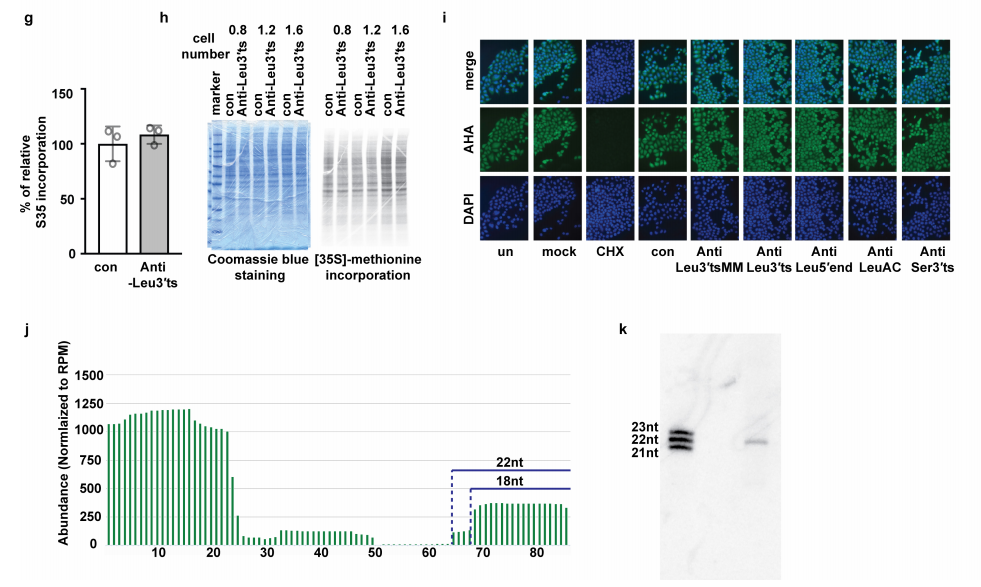
图10. 22-nt LeuCAG3‘tsRNA与细胞活性有关

图11. LeuCAG3‘tsRNA是核糖体生物发生所必需的
研究案例三
A tRNA-derived small RNA regulates ribosomal protein S28 protein levels after translation initiation in humans and mice[17]
中文题目:tRNA来源的小RNA调节人和小鼠翻译启动后核糖体蛋白S28蛋白的水平
发表时间:2019-12-17
发表期刊:Cell reports
影响因子:9.995
DOI:10.1016/j.celrep.2019.11.062
tRNA衍生的小RNA(tsRNAs)参与了许多细胞过程,然而具体的生物学机制还不是很清楚。以往研究发现Leu-CAG tRNA衍生小RNA (LeuCAG3′tsRNA)的3’端通过维持RPS28蛋白的水平来调节核糖体的生物发生。tsRNA可以结合核糖体蛋白RPS28的 mRNA,改变其二级结构并增强翻译。研究发现在灵长类动物中存在功能性的3’UTR靶点,而在许多脊椎动物中存在CDS靶点。接着,证明了tsRNA可以通过与CDS靶点相互作用来调节小鼠Rps28翻译。作者进一步证明在两个物种中mRNA翻译的变化发生在启动后的一个步骤中。本研究结果表明LeuCAG3′tsRNA可能通过一种保守的基因调控机制维持脊椎动物核糖体的生物发生。
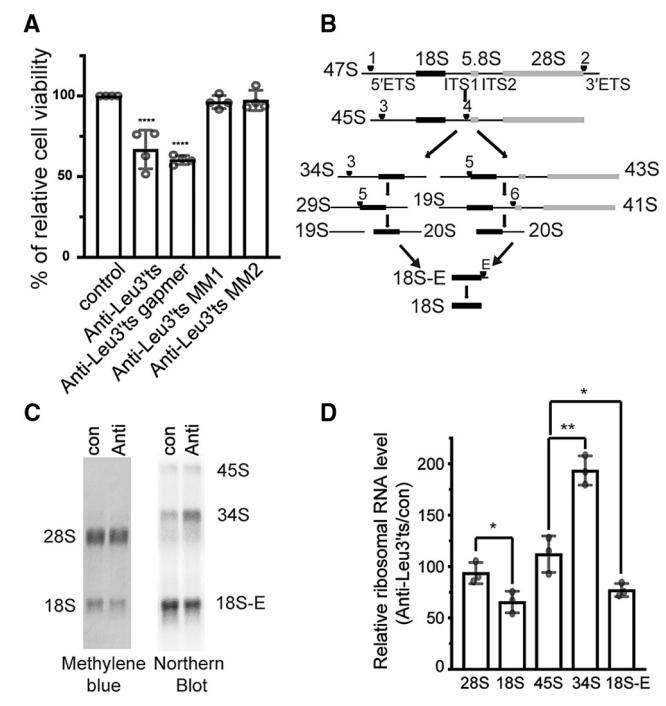
图12. LeuCAG3’ tsRNA是小鼠细胞中18S rRNA加工所必需的
研究案例四
Ribosomes guide pachytene piRNA formation on long intergenic piRNA precursors[18]
中文题目:核糖体在长链基因间piRNA 前体上介导粗线期 piRNA 的形成
发表时间:2020-02-03
发表期刊:Nature cell biology
影响因子:28.213
DOI:10.1038/s41556-019-0457-4
本研究通过翻译组测序(Ribo-seq)和小RNA测序(small RNA sequencing),发现核糖体广泛结合在piRNA前体上,并且其5´端和成熟piRNA的5´端高度重合。数据结果表明成熟piRNA的产生位置由核糖体停留在piRNA前体上的位置决定。为了探究核糖体稳定停留在长非编码RNA上的生物学机制,研究者通过分析Ribo-seq数据并利用翻译抑制剂Harringtonine以及Mov10l1和Tdrd5两种突变体,发现一些很短的上游开放阅读框存在于这些长非编码RNA的5´端。进一步研究发现在这些开放阅读框内的核糖体印记(Ribosome Footprint)呈现3nt的周期性特征,同时可以检测到蛋白表达产物,表明核糖体可以翻译这些开放阅读框。本研究首次发现粗线期piRNA的形成被核糖体介导,揭示了核糖体的又一种新功能。

图13. 粗线期piRNA前体与核糖体相关
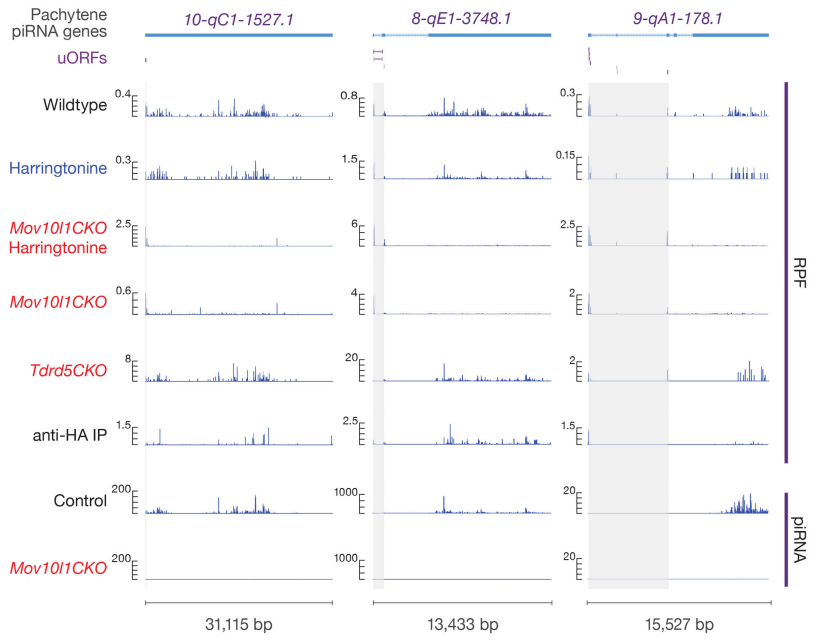
图14. 核糖体与piRNA前体结合
研究案例五
tRF-Gln-CTG-026 ameliorates liver injury by alleviating global protein synthesis[19]
中文题目:TRF-Gln-CTG-026减弱整体蛋白合成以改善肝损伤
发表时间::2023-04-03
发表期刊:Signal transduction and targeted therapy
影响因子:38.104
DOI:10.1038/s41392-023-01351-5
tsRNAs(tRNA衍生的小RNA)作为应激反应的产物,在应激反应和损伤调节中发挥着重要的作用。然而,在很大程度上仍不清楚tsRNAs是否可以改善肝脏损伤。基于此,作者利用NSUN2(NOP2/Sun结构域家族)的缺失作为tsRNAs生成模型,展示了tsRNAs在减轻肝损伤中的作用。NSUN2缺失减少了tRNAs的甲基尿苷(methyluridine-U5 ,m5U)和胞嘧啶C5(m5C),随后产生各种tsRNAs,特别是I类tsRNAs(TRF-1s)。通过进一步筛选,作者发现TRF-Gln-CTG-026(tG026)是最理想的tRF-1,它通过减弱TSR1与Pre-40S核糖体的结合来抑制总蛋白的合成,从而减轻肝损伤。这项研究表明tsRNA减少总蛋白合成在肝脏损伤和修复中的潜力,为肝脏损伤提供了一种潜在的治疗策略。
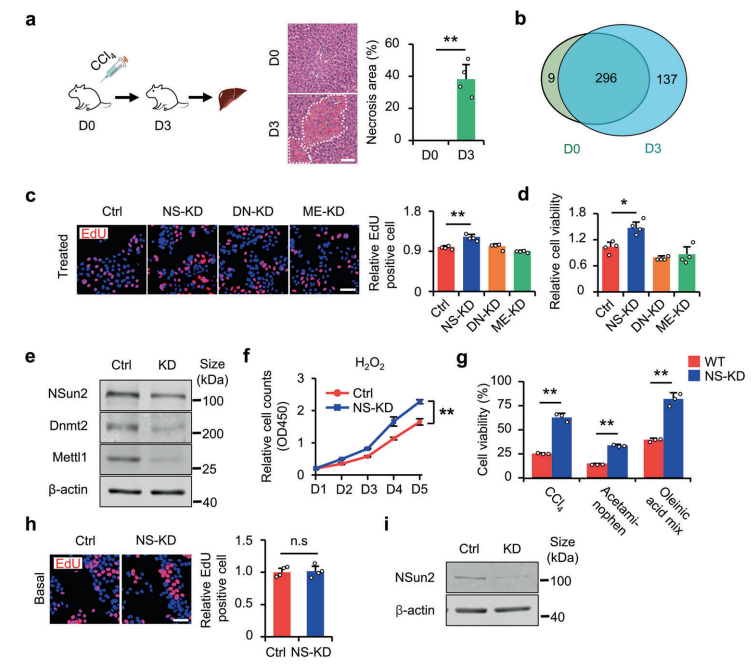
图15. NS-KD减轻细胞损伤
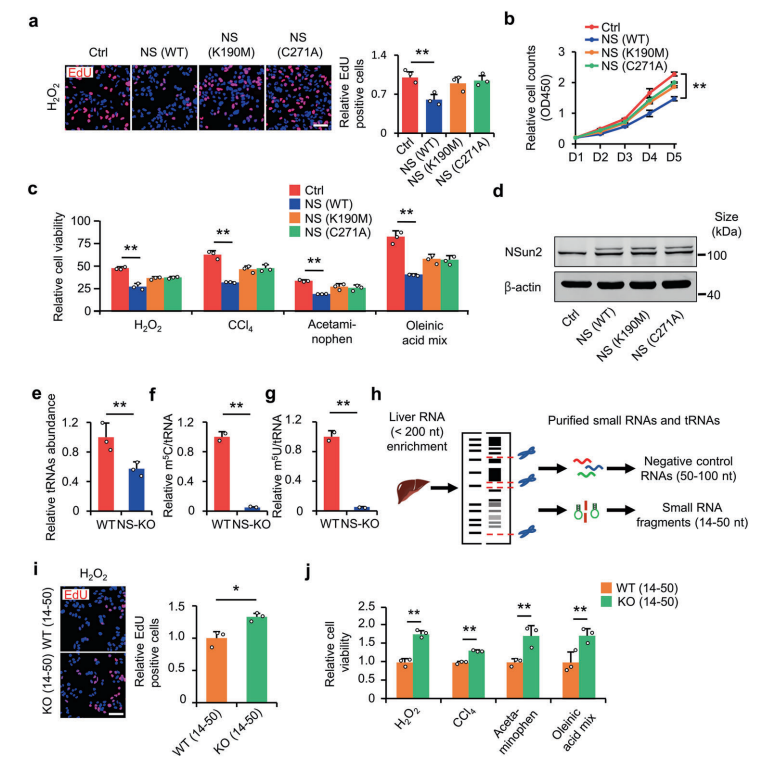
图16. NS-KO衍生的tsRNAs可减轻肝脏损伤
写在最后
液体活检,千亿蓝海:非编码小RNA,乍露锋芒。tsRNA、rsRNA等sncRNA,作为一种更有潜力的液体活检生物标志物,以往囿于其修饰干扰RNA-seg文库制备而无法检测:而现如今我们拥有了检测它们的利器PANDORA-seq,我们可以探索这片星辰大海,去挖掘更多疾病诊断、预后、疗效评估液体活检生物标志物。千亿癌症早筛市场,亟需更多的好产品,把肿瘤扼杀在摇篮中。下一个明星标志物,或许就出现在非编码小RNA中。
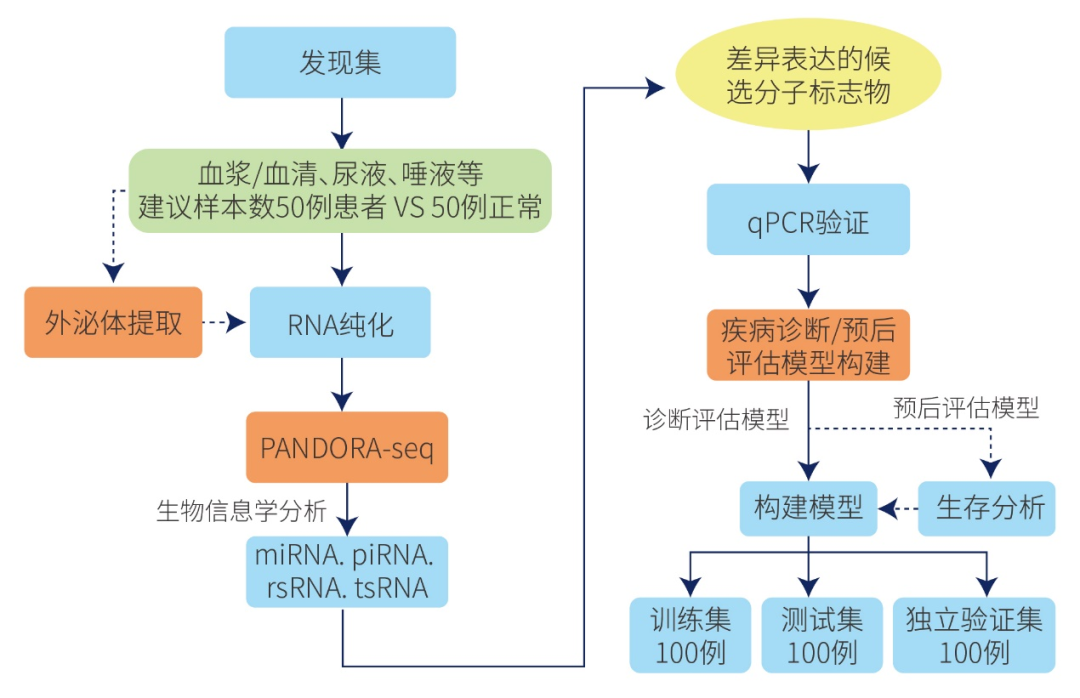
图17. PANDORA-seq进行非编码小RNA疾病标志物研究路径
广州表观生物科技有限公司在国内率先推出PANDORA-seq等测序服务,还有基于机器学习算法构建疾病诊断、预后评估模型的技术服务。如果你对这个方向也感兴趣的话,咨询我们的科研顾问,还可以领取专属福利:潘多拉测序研究方案研究资料包!
参考文献
1. Taft, R.J., et al., Non‐coding RNAs: regulators of disease. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland, 2010. 220(2): p. 126-139.
2. Hu, F., et al., tsRNA-5001a promotes proliferation of lung adenocarcinoma cells and is associated with postoperative recurrence in lung adenocarcinoma patients. Translational Lung Cancer Research, 2021. 10(10): p. 3957.
3. Cable, J., et al., Noncoding RNAs: biology and applications—a Keystone Symposia report. Annals of the New York Academy of Sciences, 2021. 1506(1): p. 118-141.
4. Hindorff, L.A., et al., Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences, 2009. 106(23): p. 9362-9367.
5. Ricano-Ponce, I. and C. Wijmenga, Mapping of immune-mediated disease genes. Annual review of genomics and human genetics, 2013. 14: p. 325-353.
6. Esteller, M., Non-coding RNAs in human disease. Nature reviews genetics, 2011. 12(12): p. 861-874.
7. Babski, J., et al., Small regulatory RNAs in archaea. RNA biology, 2014. 11(5): p. 484-493.
8. Kiss, T., Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell, 2002. 109(2): p. 145-148.
9. Lindsay, M.A., et al., Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays in biochemistry, 2013. 54: p. 53-77.
10. Tycowski, K.T., M.-D. Shu, and J.A. Steitz, A mammalian gene with introns instead of exons generating stable RNA products. Nature, 1996. 379(6564): p. 464-466.
11. Zhou, F., et al., AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nature cell biology, 2017. 19(7): p. 844-855.
12.Zhang, B., et al., Changes in snoRNA and snRNA abundance in the human, chimpanzee, macaque, and mouse brain. Genome biology and evolution, 2016. 8(3): p. 840-850.
13. Shigematsu, M. and Y. Kirino, Making invisible RNA visible: discriminative sequencing methods for RNA molecules with specific terminal formations. Biomolecules, 2022. 12(5): p. 611.
14. Shi, J., et al., PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nature cell biology, 2021. 23(4): p. 424-436.
15.Liu, J., et al., Paternal phthalate exposure-elicited offspring metabolic disorders are associated with altered sperm small RNAs in mice. Environment International, 2023. 172: p. 107769.
16. Kim, H.K., et al., A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature, 2017. 552(7683): p. 57-62.
17. Kim, H.K., et al., A tRNA-derived small RNA regulates ribosomal protein S28 protein levels after translation initiation in humans and mice. Cell reports, 2019. 29(12): p. 3816-3824. e4.
18. Sun, Y.H., et al., Ribosomes guide pachytene piRNA formation on long intergenic piRNA precursors. Nature cell biology, 2020. 22(2): p. 200-212.
19. Ying, S., et al., tRF-Gln-CTG-026 ameliorates liver injury by alleviating global protein synthesis. Signal Transduction and Targeted Therapy, 2023. 8(1): p. 144.
原文地址:https://zhuanlan.zhihu.com/p/633947793 |
|
 /3
/3 